
However, larger sample sizes come at the expense of lower mRNA detection sensitivity. Previous benchmarking studies using somatic cell lines or peripheral blood mononuclear cells (PBMCs) reported that high-throughput single-cell RNA-seq methods generally enabled broader sampling of diverse populations at a lower per-cell cost. A more complete picture of cellular transcriptional activity facilitates the identification of differentially-expressed (DE) marker genes and positively impacts the mapping of cells against reference immune cell signatures. Also, increased sensitivity in detecting individual mRNA transcripts results in more comprehensive cellular profiles, which greatly advances the characterization of immune sub-types. Methods that recover a larger fraction of cells in a cost-efficient manner benefit studies that sample tissues containing few immune cells. Efficient recovery across these diverse cell types impacts the fidelity of cell-composition analyses. Immune cells constitute a broad range of cell types across various lineages, activation states, and cell sizes. The relatively small size and low mRNA content of immune cells may impact the performance of single-cell RNA-seq methods differently than was previously described using larger cells. Mixtures of different cell sizes are particularly complex as small cells contain low total number of transcripts and therefore, are difficult to distinguish from ambient noise. Several key factors, such as variable capture and amplification efficiencies during library preparation, impact the ability of single-cell RNA-seq techniques to accurately and comprehensively characterize immune-cell diversity. As recent advances have increased cell throughput and lowered per-cell costs, the number of high-throughput single-cell RNA-seq techniques that can process more than a thousand cells per experiment has increased. These single-cell technologies have enabled immunologists to characterize inflammation and immune responses to cancer, uncovering previously uncharacterized cellular diversity and cell-type specific transcriptional responses. Although techniques such as FACS and mass cytometry are useful for studying cellular diversity according to well-characterized cell-surface-protein markers, the advent of single-cell RNA sequencing (RNA-seq) has expanded the power to characterize individual immune cells from a defined set of cell-surface markers to the entire transcriptome for last few years. Understanding the cellular diversity underlying immune responses is an important component of immunological research. Overall, our characterization of immune cell mixtures provides useful metrics, which can guide selection of a high-throughput single-cell RNA-seq method for profiling more complex immune-cell heterogeneity usually found in vivo. We demonstrate that these methods have fewer dropout events, which facilitates the identification of differentially-expressed genes and improves the concordance of single-cell profiles to immune bulk RNA-seq signatures. We observed higher mRNA detection sensitivity with the 10x Genomics 5′ v1 and 3′ v3 methods.
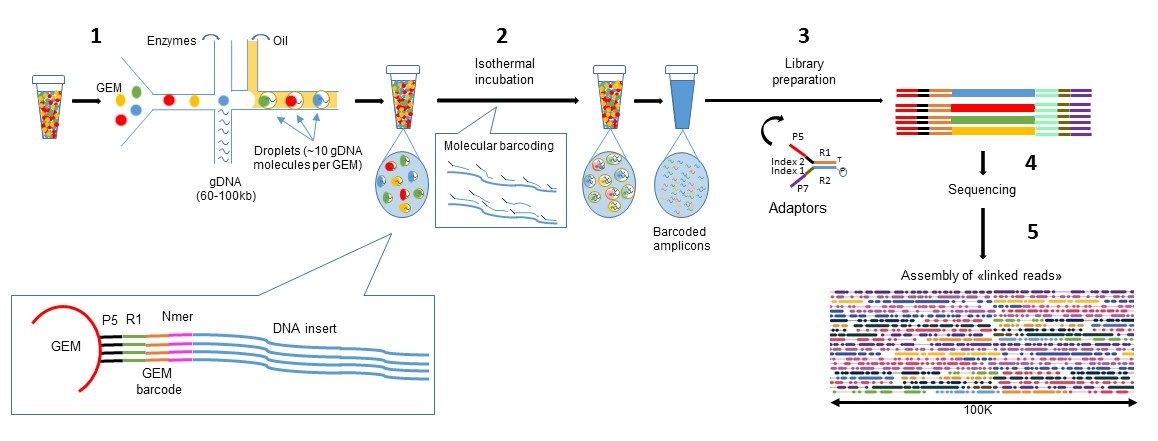
We evaluated methods by their cell recovery rate, library efficiency, sensitivity, and ability to recover expression signatures for each cell type. We prepared 21 libraries under identical conditions of a defined mixture of two human and two murine lymphocyte cell lines, simulating heterogeneity across immune-cell types and cell sizes. Here, we systematically benchmarked seven high-throughput single-cell RNA-seq methods. This issue is often compounded by limited sample availability and limited prior knowledge of heterogeneity, which can confound data interpretation.

However, single-cell methods inherently suffer from limitations in the recovery of complete transcriptomes due to the prevalence of cellular and transcriptional dropout events. Elucidation of immune populations with single-cell RNA-seq has greatly benefited the field of immunology by deepening the characterization of immune heterogeneity and leading to the discovery of new subtypes.


 0 kommentar(er)
0 kommentar(er)
